## Enhanced Classifier ###
# Load packages
devtools::source_url("https://github.com/r2j2ritson/RSpatial_Wildlife/blob/main/R/coords_as_sf.R?raw=TRUE")
devtools::source_url("https://github.com/r2j2ritson/RSpatial_Wildlife/blob/main/R/instaload.R?raw=TRUE")
instaload(c('data.table','lubridate','plyr','zoo','mapboxer','rmarkdown','markdown','tinytex',
'ggplot2','ggmap','dplyr'))
#### USER INPUT - set up directories; create file copy to overwrite classifications
species <- "mule deer"
inpath <- "K:/Wildlife/sbergen/forRitson/nsd/ID_MD_2019_NSD"
scriptdir <- "C:/Users/rritson/Documents/Projects/NSD/sandbox"
setwd(inpath) # change working directory to deer or elk folder
i=5
herd <- paste0("Sel",i)
file <- paste0("ID_MD_2019_locs_cleaned_NSD_sel",i,"_go.csv")
rawdir <- paste0(getwd(), "/unclassified_data")
datadir <- paste0(getwd(), "/classified_data")
outfile <- paste0(datadir, "/", file)
###
# Read in the file prepared in last script
all_recs <- outfile %>%
fread(., header = TRUE, stringsAsFactors = FALSE) %>%
dplyr::rowwise(.) %>%
dplyr::mutate(GMT = as.POSIXct(paste0(Year,"-",Month,"-",Day," ",Hour,":",Minute,":",Second),
format = "%Y-%m-%d %H:%M:%S", tz = "GMT")) %>%
dplyr::ungroup(.) %>%
as.data.frame(.)
str(all_recs)
yrs <- unique(year(all_recs$GMT))
yrs <- sort(yrs)
#######################
savecounter = 0
# Prep Color Palette
hab_palette <- data.frame("Habitat" = c("Discard",
"Not assigned",
"Not assigned",
"blank",
"Winter",
"Winter",
"Summer",
"Summer",
"SpringMig",
"FallMig",
"Resident",
"Nomad"),
"color" = c("#BEBEBE",
"#000000",
"#000000",
"#000000",
"#0000FF",
"#BFEFFF",
"#FFFF00",
"#EEDD82",
"#B8860B",
"#008000",
"#FF00FF",
"#40E0D0"),
"Hab_code" =c("Discard",
"not assigned",
NA,
"",
"winter",
"wntmov",
"summer",
"summov",
"fallmig",
"sprmig",
"Resident",
"Nomad"))
# Classify Dates Function
classify_dates <- function(id_recs){
## Look-up Tables
spring_lut <- id_recs %>%
dplyr::filter(Hab_code == "sprmig") %>%
dplyr::group_by(habitat) %>%
dplyr::summarise(Start = min(GMT),
End = max(GMT)) %>%
dplyr::arrange(.,Start)
fall_lut <- id_recs %>%
dplyr::filter(Hab_code == "fallmig") %>%
dplyr::group_by(habitat) %>%
dplyr::summarise(Start = min(GMT),
End = max(GMT)) %>%
dplyr::arrange(.,Start)
## Func Codes
last_summer <- function(){
paste0("summer", (as.numeric(year)-1))
}
last_winter <- function(){
paste0("winter", (as.numeric(year)-1))
}
next_summer <- function(){
paste0("summer", (as.numeric(year)+1))
}
next_winter <- function(){
paste0("winter", (as.numeric(year)+1))
}
current_summer <- function(){
paste0("summer", year)
}
current_winter <- function(){
paste0("winter", year)
}
## Classify Dates
### Only One Spring Migration classified
if(nrow(spring_lut) == 1 & nrow(fall_lut) == 0){ #classic partial sequence
id_recs$habitat <- ifelse(id_recs$color == "#000000" & id_recs$GMT < spring_lut$Start,
current_winter(), id_recs$habitat)
id_recs$habitat <- ifelse(id_recs$color == "#000000" & id_recs$GMT > spring_lut$End,
current_summer(), id_recs$habitat)
}
### Only One Fall Migration classified
if(nrow(spring_lut) == 0 & nrow(fall_lut) == 1){
id_recs$habitat <- ifelse(id_recs$color == "#000000" & id_recs$GMT < fall_lut$Start,
last_summer(), id_recs$habitat)
id_recs$habitat <- ifelse(id_recs$color == "#000000" & id_recs$GMT > fall_lut$End,
current_winter(), id_recs$habitat)
}
### One Spring and One Fall Migration classified
if(nrow(spring_lut) == 1 & nrow(fall_lut) == 1){
if(spring_lut$Start < fall_lut$Start){ #Classic complete sequence
id_recs$habitat <- ifelse(id_recs$color == "#000000" & id_recs$GMT < spring_lut$Start,
current_winter(), id_recs$habitat)
id_recs$habitat <- ifelse(id_recs$color == "#000000" & id_recs$GMT > spring_lut$End & id_recs$GMT < fall_lut$Start,
current_summer(), id_recs$habitat)
id_recs$habitat <- ifelse(id_recs$color == "#000000" & id_recs$GMT > fall_lut$End,
next_winter(), id_recs$habitat)
}else{
id_recs$habitat <- ifelse(id_recs$color == "#000000" & id_recs$GMT < fall_lut$Start,
last_summer(), id_recs$habitat)
id_recs$habitat <- ifelse(id_recs$color == "#000000" & id_recs$GMT > fall_lut$End & id_recs$GMT < spring_lut$Start,
current_winter(), id_recs$habitat)
id_recs$habitat <- ifelse(id_recs$color == "#000000" & id_recs$GMT > spring_lut$End,
current_summer(), id_recs$habitat)
}
}
### Two Fall Migrations and One Spring Spring Migration classified
if(nrow(spring_lut) == 1 & nrow(fall_lut) == 2){
id_recs$habitat <- ifelse(id_recs$color == "#000000" & id_recs$GMT < fall_lut$Start[1],
last_summer(), id_recs$habitat)
id_recs$habitat <- ifelse(id_recs$color == "#000000" & id_recs$GMT < spring_lut$Start & id_recs$GMT > fall_lut$End[1],
current_winter(), id_recs$habitat)
id_recs$habitat <- ifelse(id_recs$color == "#000000" & id_recs$GMT > spring_lut$End & id_recs$GMT < fall_lut$Start[2],
current_summer(), id_recs$habitat)
id_recs$habitat <- ifelse(id_recs$color == "#000000" & id_recs$GMT > fall_lut$End[2],
next_winter(), id_recs$habitat)
}
### One Fall Migrations and Two Spring Spring Migration classified
if(nrow(spring_lut) == 2 & nrow(fall_lut) == 1){
id_recs$habitat <- ifelse(id_recs$color == "#000000" & id_recs$GMT < spring_lut$Start[1],
current_winter(), id_recs$habitat)
id_recs$habitat <- ifelse(id_recs$color == "#000000" & id_recs$GMT > spring_lut$End[1] & id_recs$GMT < fall_lut$Start,
current_summer(), id_recs$habitat)
id_recs$habitat <- ifelse(id_recs$color == "#000000" & id_recs$GMT > fall_lut$End & id_recs$GMT < spring_lut$Start[2],
next_winter(), id_recs$habitat)
id_recs$habitat <- ifelse(id_recs$color == "#000000" & id_recs$GMT > spring_lut$End[2],
next_summer(), id_recs$habitat)
}
### Add Years to Winter and Summer Movements
id_recs$Hab_code <- stringr::str_split_fixed(id_recs$habitat,"[:digit:]",2)[,1]
winter_lut <- id_recs %>%
dplyr::filter(Hab_code == "winter") %>%
dplyr::group_by(habitat) %>%
dplyr::summarise(Start = min(GMT),
End = max(GMT)) %>%
dplyr::arrange(.,Start) %>%
dplyr::rowwise(.) %>%
dplyr::mutate(Year = stringr::str_split_fixed(habitat,"winter",2)[,2]) %>%
dplyr::ungroup(.) %>%
as.data.frame(.)
summer_lut <- id_recs %>%
dplyr::filter(Hab_code == "summer") %>%
dplyr::group_by(habitat) %>%
dplyr::summarise(Start = min(GMT),
End = max(GMT)) %>%
dplyr::arrange(.,Start) %>%
dplyr::rowwise(.) %>%
dplyr::mutate(Year = stringr::str_split_fixed(habitat,"summer",2)[,2]) %>%
dplyr::ungroup(.) %>%
as.data.frame(.)
if(nrow(winter_lut) >=1){
for(i in 1:nrow(winter_lut)){
id_recs$habitat <- ifelse(id_recs$habitat == "wntmov" & id_recs$GMT > winter_lut$Start[i] & id_recs$GMT < winter_lut$End[i],
paste0("wntmov", winter_lut$Year[i]), id_recs$habitat)
}
}
if(nrow(summer_lut) >=1){
for(i in 1:nrow(summer_lut)){
id_recs$habitat <- ifelse(id_recs$habitat == "summov" & id_recs$GMT > summer_lut$Start[i] & id_recs$GMT < summer_lut$End[i],
paste0("summov", winter_lut$Year[i]), id_recs$habitat)
}
}
id_recs$Hab_code <- NULL
id_recs$color <- NULL
id_recs$Hab_code <- stringr::str_split_fixed(id_recs$habitat,"[:digit:]",2)[,1]
id_recs <- dplyr::left_join(id_recs,hab_palette[,c(-1)],by="Hab_code")
return(id_recs)
}
# Begin Year loop
for(y in 1:length(yrs)){
print(paste("Looking for year", yrs[y], "records needing classification"))
## Set-up ###
# Subset dataset yr by yr; Jan-Feb of next yr kept too in case fall migration carries over
yr_recs <- all_recs[year(all_recs$GMT) == yrs[y] |
year(all_recs$GMT) == yrs[y] + 1 &
month(all_recs$GMT) < 5, ]
# Read in overall notes table
notes_tab <- read.csv(paste0(getwd(), "/unsorted_output/", herd, "/", yrs[y],
"/", yrs[y], "_overall.csv"), header = TRUE)
notes_tab[] <- lapply(notes_tab, as.character)
## Create list of individuals to analyze ###
if(exists("id_list")){
if(! is.null(id_list)){
individs_list <- id_list
} else {
fb <- readline("Pick-up where you left off (pu) OR review all (ra)? ")
if(fb == "pu"){
individs_list <- notes_tab[is.na(notes_tab$Skip) | notes_tab$Skip == "", ]$Animal_ID
}else{
individs_list <- notes_tab$Animal_ID
}
}
} else {
fb <- readline("Pick-up where you left off (pu) OR review all (ra)? ")
if(fb == "pu"){
individs_list <- notes_tab[is.na(notes_tab$Skip) | notes_tab$Skip == "", ]$Animal_ID
}else{
individs_list <- notes_tab$Animal_ID
}
}
individs_list <- individs_list[which(individs_list %in% all_recs$Animal_ID)]
# If empty, review revisits?
if(length(individs_list) == 0){
revisit <- readline("No individuals left to be classified. Review revisits? Y or N ")
if(revisit == "Y"){
individs_list <- notes_tab[notes_tab$Skip == "N" & notes_tab$Revisit == "Y", ]$Animal_ID
individs_list <- individs_list[which(individs_list %in% all_recs$Animal_ID)]
}
}
if(length(individs_list)==0){
message("There are no revisits remaining in this file.")
next
}
## Begin Migration Analysis ###
# Individual loop
for(id in 1:length(individs_list)){
# Check previous plots if revisiting...
if(revisit == "Y"){
## Query previous NSDs
source("K:/Wildlife/sbergen/forRitson/nsd/ID_MD_2019_NSD/query_previous_nsds.R")
find_nsds(id = individs_list[id], species = species)
## What are we checking?
if(notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Skip == "Y"){
message("This individual was previously skipped.")
}else if(notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Strategy == "M"){
message("This individual was previously labeled a migrant.")
}else if(notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Strategy == "MM"){
message("This individual was previously labeled a mixed-migrant.")
}else if(notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Strategy == "R"){
message("This individual was previously labeled a resident.")
}else if(notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Strategy == "N"){
message("This individual was previously labeled a nomad.")
}else if(notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Strategy == "U"){
message("This individual was previously labeled unknown.")
}
## View current NSD plot
message(paste("Check:",notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Notes))
shell.exec(paste0(getwd(), "/unsorted_output/",herd, "/", yrs[y],"/",individs_list[id], "_graphic.html", sep = ""))
## Resume?
skip_revisit <- readline("Keep current NSD (skip this revisit)? Y or N ")
notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Revisit <- readline("Revisit another time? Y or N ")
if(skip_revisit == "Y"){
fwrite(notes_tab, paste0(getwd(), "/unsorted_output/", herd, "/", yrs[y],
"/", yrs[y], "_overall.csv"), row.names = FALSE,
dateTimeAs = c("write.csv"))
system2("taskkill",args="/im chrome.exe")
next
}
}
# Examine Movements...
repeat{
id_recs <- yr_recs[yr_recs$Animal_ID == individs_list[id], ]
#Show locs
print({
shp <- coords_as_sf(id_recs,"WGS84_Long","WGS84_Lat","WGS84")
tile1 <- maptiles::get_tiles(shp, provider = "Esri.WorldTopoMap", crop=T, zoom=10, cachedir = ".") %>%
terra::project(.,terra::crs(shp))
grDevices::x11()
ggplot() +
tidyterra::geom_spatraster_rgb(data = tile1, maxcell = Inf) +
geom_sf(data = shp) +
labs(title = paste0("Animal ID: ",individs_list[id]))
})
unlink(paste0(inpath,"/EsriWTM"),recursive = T)
## Step 1: Select Anchor Date
repeat{
def <- readline("Use Default Anchor Date (March 1) or reveiw options? Y or N ")
if(def == "N"){
rmarkdown::render(paste0(scriptdir, "/", "mapbox_anchor.Rmd"),
output_file = paste0(individs_list[id], "_anchors.html"),
output_dir = paste0(getwd(), "/unsorted_output/",
herd, "/", yrs[y], sep = ""))
shell.exec(paste0(getwd(), "/unsorted_output/",herd, "/", yrs[y],"/",individs_list[id], "_anchors.html", sep = ""))
repeat{
anc_date <- readline("Enter Anchor Date (M-DD) ")
print(paste("Using Anchor Date ", anc_date))
orig_date <- as.POSIXct(strptime(paste0(yrs[y], "-",anc_date," 00:00:00"), format = "%Y-%m-%d %H:%M:%S"),
origin = "1970-01-01 00:00:00")
orig_date_recs <- id_recs[abs(id_recs$GMT - orig_date) ==
min(abs(id_recs$GMT - orig_date)), ][1, ]
print(paste("NSD calculated from origin at", orig_date_recs$GMT))
id_recs[year(id_recs$GMT) == yrs[y] , "Root_NSD"] <-
((id_recs[year(id_recs$GMT) == yrs[y] , "IDTM_X"] - orig_date_recs$IDTM_X)^2 +
(id_recs[year(id_recs$GMT) == yrs[y] , "IDTM_Y"] - orig_date_recs$IDTM_Y)^2)^0.5
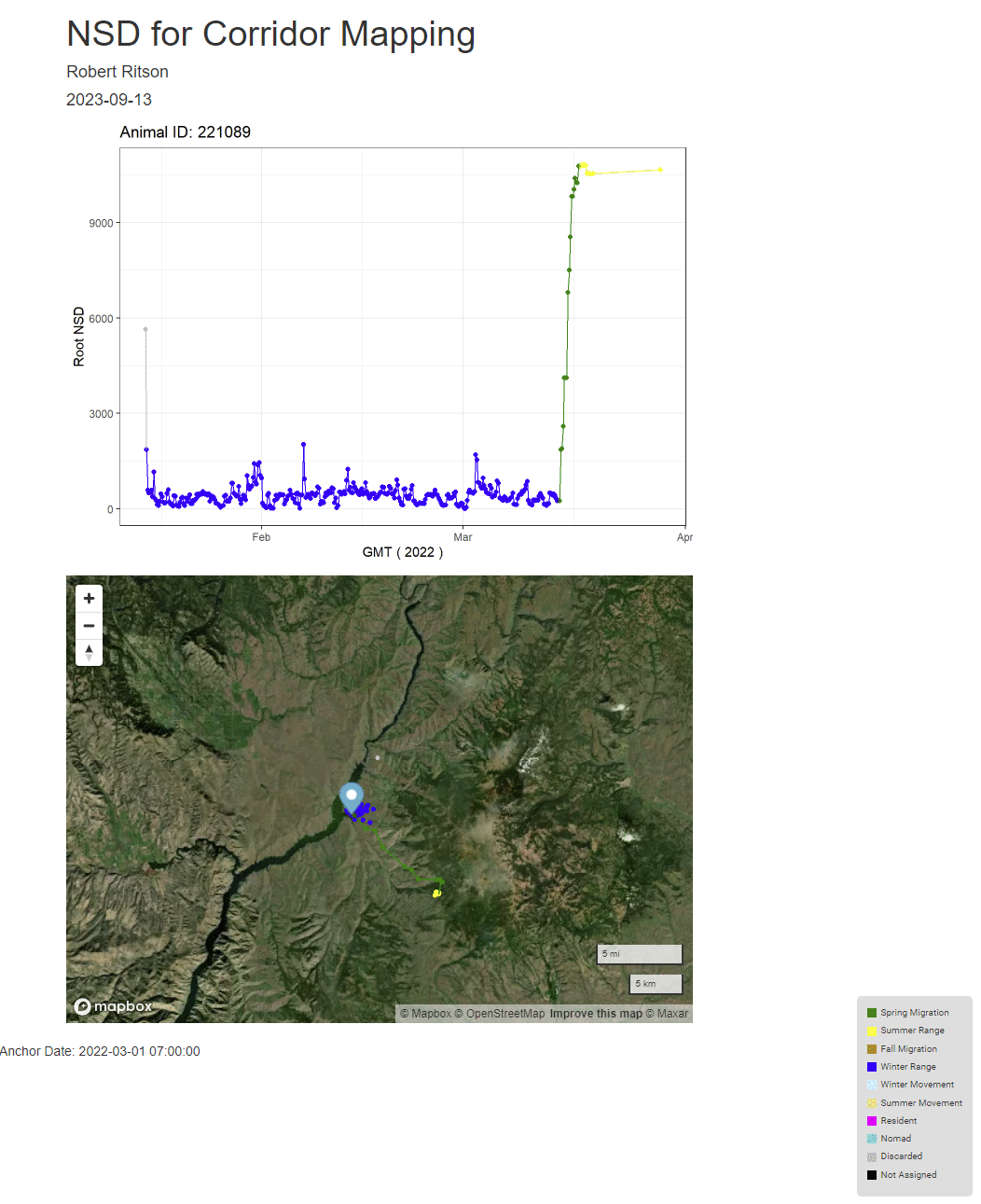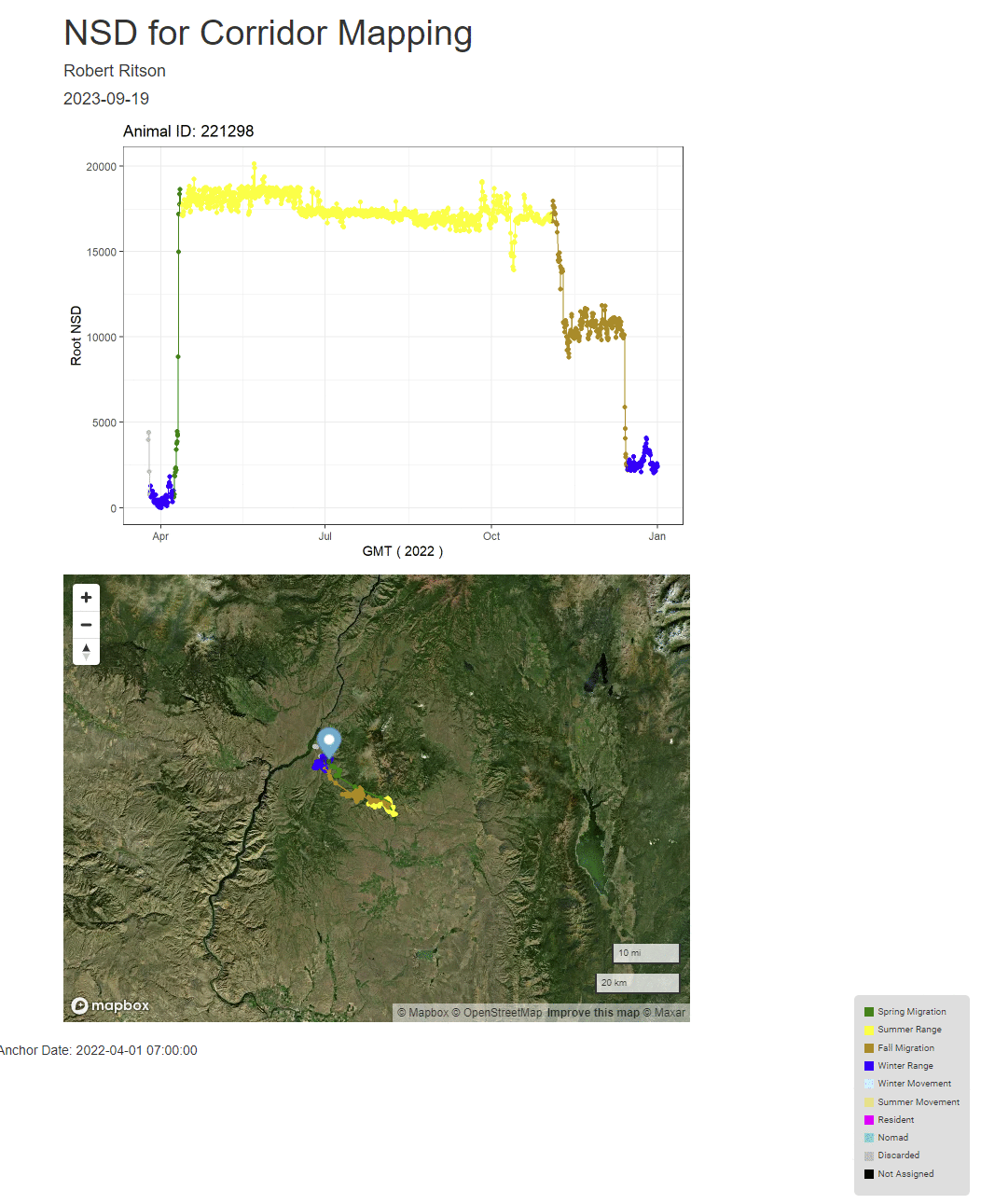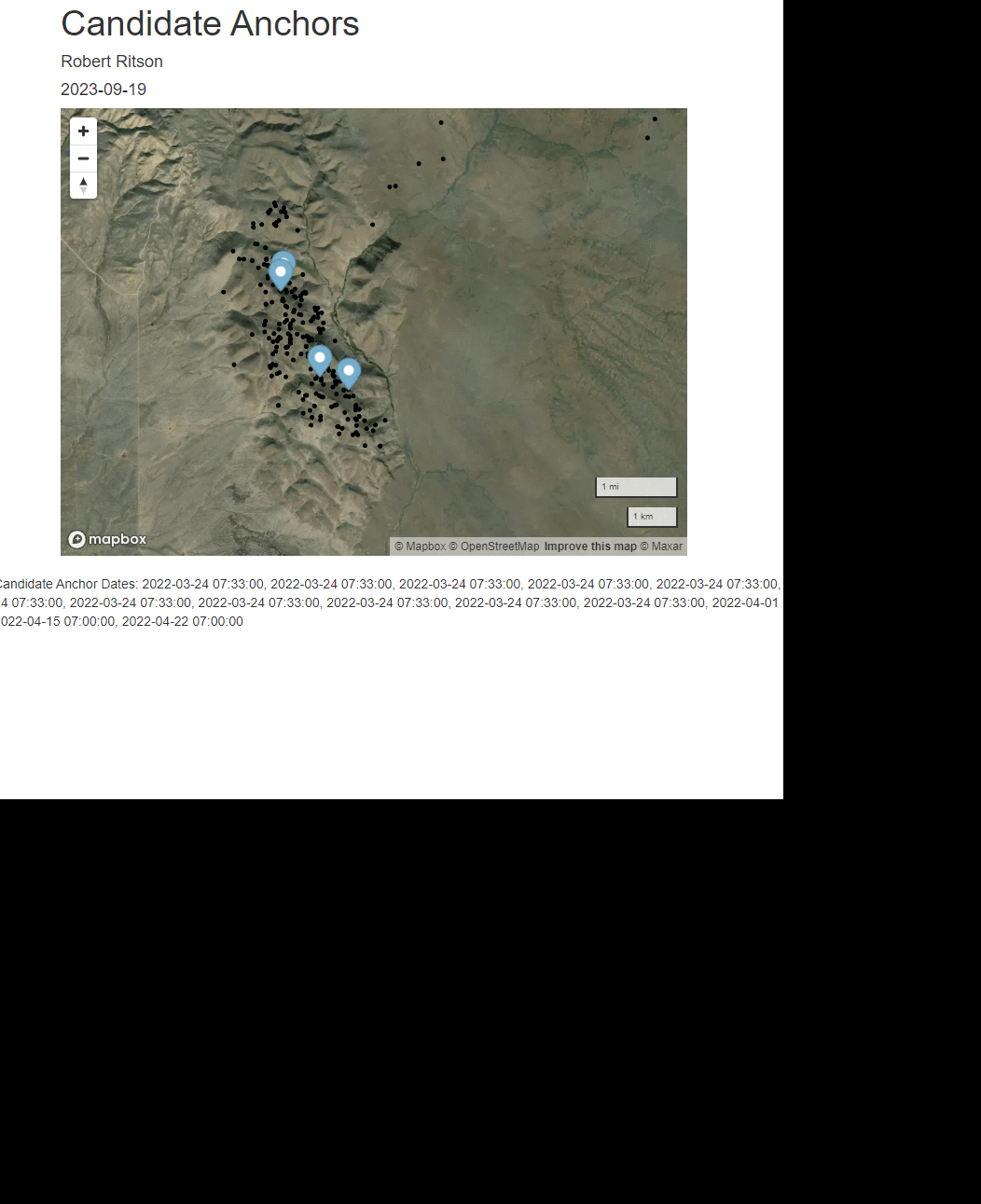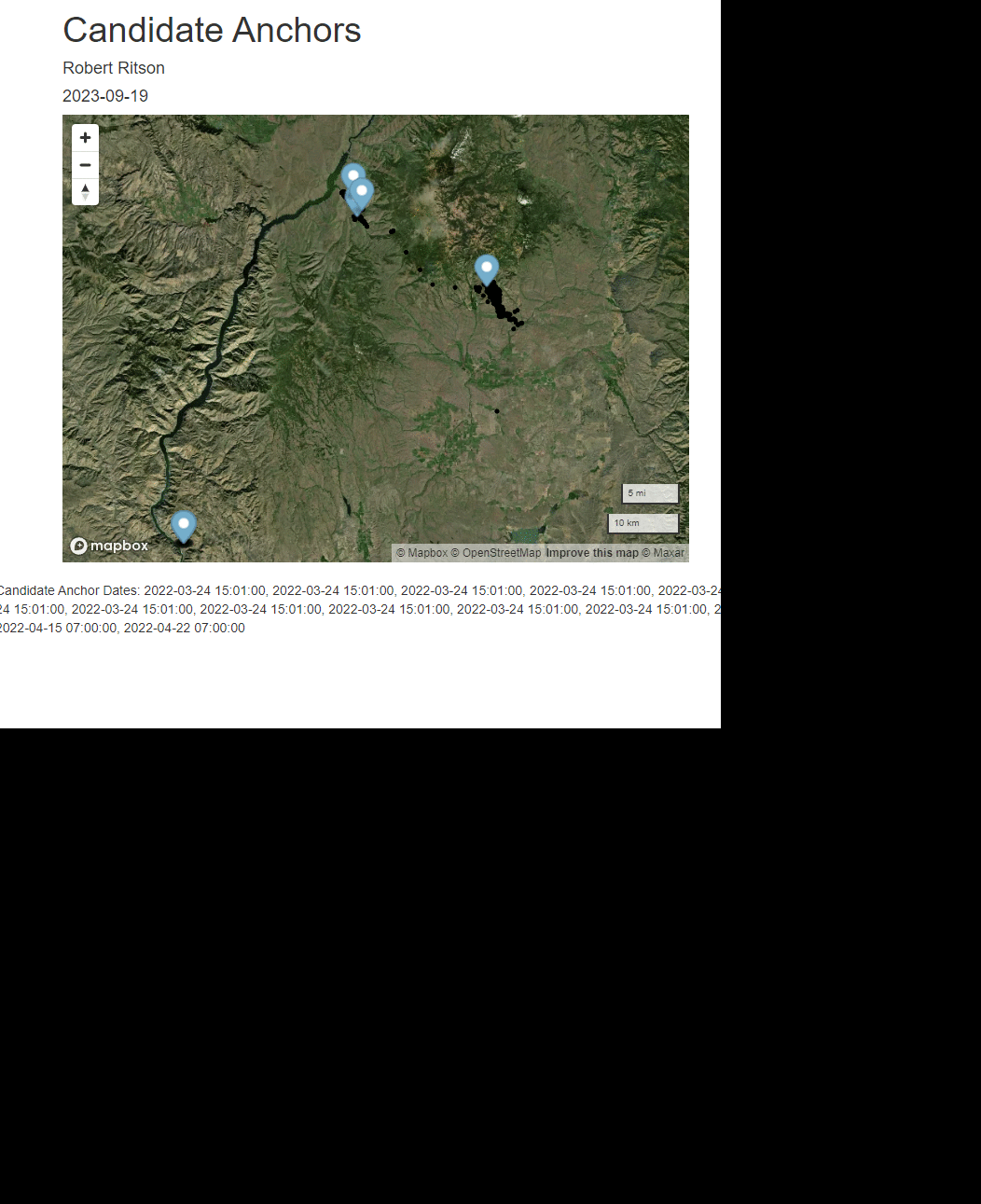
print(ggplot(id_recs,aes(x=GMT,y=Root_NSD)) +
geom_point(color=id_recs$color) + geom_line(color=id_recs$color) +
labs(x="GMT",y="Root_NSD",
title = paste0("Animal ID: ",individs_list[id]),
caption=paste0("Anchor Date: ",format.Date(orig_date,"%B %d"))))
BR <- readline("NSD and Anchor Date look good? Y or N ")
if(BR == "Y"){
break
}
}
}else{
# Set date of origin
print("Using Default Anchor Date (March 1).")
orig_date <- as.POSIXct(strptime(paste0(yrs[y], "-3-01 00:00:00"), format = "%Y-%m-%d %H:%M:%S"),
origin = "1970-01-01 00:00:00") # origin date is March 1
orig_date_recs <- id_recs[abs(id_recs$GMT - orig_date) ==
min(abs(id_recs$GMT - orig_date)), ][1, ]
print(paste("NSD calculated from origin at", orig_date_recs$GMT))
}
notes_tab[notes_tab$Animal_ID == individs_list[id], ]$AnchorDate <- format.Date(orig_date,"%B %d")
id_recs[year(id_recs$GMT) == yrs[y] , "Root_NSD"] <-
((id_recs[year(id_recs$GMT) == yrs[y] , "IDTM_X"] - orig_date_recs$IDTM_X)^2 +
(id_recs[year(id_recs$GMT) == yrs[y] , "IDTM_Y"] - orig_date_recs$IDTM_Y)^2)^0.5
print(ggplot(id_recs,aes(x=GMT,y=Root_NSD)) +
geom_point(color=id_recs$color) + geom_line(color=id_recs$color) +
labs(x="GMT",y="Root_NSD",
title = paste0("Animal ID: ",individs_list[id]),
caption=paste0("Anchor Date: ",format.Date(orig_date,"%B %d"))))
BR <- readline("NSD and Anchor Date look good? Y or N ")
if(BR == "Y"){
break
}
}
###
# Skip animal or classify migration dates
notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Skip <- readline("Skip this animal? Y or N ")
## Step 2: Trim or Discard spurious points
if(notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Skip == "N"){
notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Trim <- readline("Trim points from the graph? Y or N ")
if(notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Trim == "Y"){
discard_all <- NULL
repeat{
TR <- readline("Discard points from the top(T), bottom(B) right(R), or left(L) of graph? T, B, R, or L ")
if(TR == "T"){
print("Click below the points to discard.")
discard <- ggmap::gglocator(n=1,mercator = F)
discarded <- id_recs[as.numeric(na.omit(id_recs$Root_NSD)) > discard$Root_NSD, ]
id_recs <- id_recs[as.numeric(na.omit(id_recs$Root_NSD)) < discard$Root_NSD, ]
notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Trim_type <- ifelse(is.na(notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Trim_type),TR,
paste0(notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Trim_type,TR))
} else if(TR == "R") {
print("Click left of the points to discard.")
discard <- ggmap::gglocator(n=1,mercator = F)
discarded <- id_recs[as.numeric(na.omit(id_recs$GMT)) > discard$GMT, ]
id_recs <- id_recs[as.numeric(na.omit(id_recs$GMT)) < discard$GMT, ]
notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Trim_type <- ifelse(is.na(notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Trim_type),TR,
paste0(notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Trim_type,TR))
} else if(TR == "L") {
print("Click right of the points to discard.")
discard <- ggmap::gglocator(n=1,mercator = F)
discarded <- id_recs[as.numeric(na.omit(id_recs$GMT)) < discard$GMT, ]
id_recs <- id_recs[as.numeric(na.omit(id_recs$GMT)) > discard$GMT, ]
notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Trim_type <- ifelse(is.na(notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Trim_type),TR,
paste0(notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Trim_type,TR))
} else if(TR == "B") {
print("Click above the points to discard.")
discard <- ggmap::gglocator(n=1,mercator = F)
discarded <- id_recs[as.numeric(na.omit(id_recs$Root_NSD)) < discard$Root_NSD, ]
id_recs <- id_recs[as.numeric(na.omit(id_recs$Root_NSD)) > discard$Root_NSD, ]
notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Trim_type <- ifelse(is.na(notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Trim_type),TR,
paste0(notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Trim_type,TR))
}
discard_all <- rbind(discard_all,discarded)
BR <- readline("Discard more points? Y or N ")
if(BR == "N"){
break
}
}
}
###
repeat{
print(ggplot(id_recs,aes(x=GMT,y=Root_NSD)) +
geom_point(color=id_recs$color) + geom_line(color=id_recs$color) +
labs(x="GMT",y="Root_NSD",
title = paste0("Animal ID: ",individs_list[id]),
caption=paste0("Anchor Date: ",format.Date(orig_date,"%B %d"))))
notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Strategy <- readline("Identify tentative migration strategy.
Please list M (migratory), MM(mixed-migratory), R(resident), N(nomadic), or U(unclear) ")
if(notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Strategy == "R"){
id_recs$habitat <- "Resident"
id_recs$Hab_code <- "Resident"
id_recs <- dplyr::left_join(id_recs[,-which(stringr::str_detect(colnames(id_recs),"color")==T)],hab_palette[,c(-1)],by="Hab_code")
rmarkdown::render(paste0(scriptdir, "/", "mapbox_plot.Rmd"),
output_file = paste0(individs_list[id], "_graphic.html"),
output_dir = paste0(getwd(), "/unsorted_output/",
herd, "/", yrs[y], sep = ""))
shell.exec(paste0(getwd(), "/unsorted_output/",herd, "/", yrs[y],"/",individs_list[id], "_graphic.html", sep = ""))
if(readline("Open the output file. Look good? Y or N ") == "Y"){
break
}else{
id_recs$habitat <- "not assigned"
id_recs$color <- "#000000"
id_recs$Hab_code <- NULL
}
}
if(notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Strategy == "N"){
id_recs$habitat <- "Nomad"
id_recs$Hab_code <- "Nomad"
id_recs <- dplyr::left_join(id_recs[,-which(stringr::str_detect(colnames(id_recs),"color")==T)],hab_palette[,c(-1)],by="Hab_code")
rmarkdown::render(paste0(scriptdir, "/", "mapbox_plot.Rmd"),
output_file = paste0(individs_list[id], "_graphic.html"),
output_dir = paste0(getwd(), "/unsorted_output/",
herd, "/", yrs[y], sep = ""))
shell.exec(paste0(getwd(), "/unsorted_output/",herd, "/", yrs[y],"/",individs_list[id], "_graphic.html", sep = ""))
if(readline("Open the output file. Look good? Y or N ") == "Y"){
break
}else{
id_recs$habitat <- "not assigned"
id_recs$color <- "#000000"
id_recs$Hab_code <- NULL
}
}
if(notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Strategy != "R" &
notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Strategy != "N"){
## Step 3: Classify Movement Dates
repeat{
# Plot NSD
print(plt <-ggplot(id_recs,aes(x=GMT,y=Root_NSD)) +
geom_point(color=id_recs$color) + geom_line(color=id_recs$color) +
labs(x="GMT",y="Root NSD",title = paste0("Animal ID: ",individs_list[id]))+
theme_bw())
mig <- readline("What kind of movement is this? Spring(sp), Fall(fa), Winter movement (wm), or Summer movement (sm)? ")
if(mig == "sp"){
print("Select Spring Migration Dates")
repeat{
# Plot NSD
print(
plt <-ggplot(id_recs,aes(x=GMT,y=Root_NSD)) +
geom_point(color=id_recs$color) + geom_line(color=id_recs$color) +
labs(x="GMT",y="Root NSD",title = paste0("Animal ID: ",individs_list[id]))+
theme_bw()
)
# Zoom in on dates of spring migration
print("Click the earlier date of the range you'd like to zoom to")
windowstart <- ggmap::gglocator(n=1,mercator = F)$GMT
print(plt + geom_vline(xintercept = windowstart))
print("Click the later date of the range you'd like to zoom to")
windowend <- ggmap::gglocator(n=1,mercator = F)$GMT
window_recs <- id_recs[as.numeric(na.omit(id_recs$GMT)) >
windowstart & as.numeric(na.omit(id_recs$GMT)) < windowend, ]
print(plt <- ggplot(window_recs,aes(x=GMT,y=Root_NSD)) +
geom_point(color=window_recs$color) + geom_line(color=window_recs$color) +
labs(x="GMT",y="Root NSD",title = paste0("Animal ID: ",individs_list[id])))
# Pick start/end dates with locator; assign/paste makes new variables for the loop
print("Click the start of the spring migration")
start <- ggmap::gglocator(n=1,mercator = F)$GMT
print(plt <- plt + geom_vline(xintercept = start, color='limegreen'))
print("Click the end of the spring migration")
end <- ggmap::gglocator(n=1,mercator = F)$GMT
print(plt <- plt + geom_vline(xintercept = end, color='darkred'))
# Identify dates in selected range
year <- substr(yrs[y], 3, 4)
spring_start <- as.POSIXct(start, origin = "1970-01-01 00:00:00",tz='GMT')
spring_end <- as.POSIXct(end, origin = "1970-01-01 00:00:00",tz='GMT')
if(spring_start > orig_date){ #spring identified in current year sequence
id_recs$habitat <- ifelse(spring_start <= id_recs$GMT & id_recs$GMT <= spring_end,paste0("sprmig", year),
id_recs$habitat)
}else{#spring identified in last year sequence
id_recs$habitat <- ifelse(spring_start <= id_recs$GMT & id_recs$GMT <= spring_end,paste0("sprmig", (as.numeric(year)-1)),
id_recs$habitat)
}
id_recs$Hab_code <- stringr::str_split_fixed(id_recs$habitat,"[:digit:]",2)[,1]
id_recs <- dplyr::left_join(id_recs[,-which(stringr::str_detect(colnames(id_recs),"color")==T)],hab_palette[,c(-1)],by="Hab_code")
# ~~*plotting magic*~~
rmarkdown::render(paste0(scriptdir, "/", "mapbox_plot.Rmd"),
output_file = paste0(individs_list[id], "_graphic.html"),
output_dir = paste0(getwd(), "/unsorted_output/",
herd, "/", yrs[y], sep = ""))
shell.exec(paste0(getwd(), "/unsorted_output/",herd, "/", yrs[y],"/",individs_list[id], "_graphic.html", sep = ""))
if(readline("Open the output file. Look good? Y or N ") == "Y"){
break
}else{
id_recs$habitat <- ifelse(spring_start <= id_recs$GMT & id_recs$GMT <= spring_end,"not assigned",
id_recs$habitat)
id_recs$color <- ifelse(spring_start <= id_recs$GMT & id_recs$GMT <= spring_end,"#000000",
id_recs$color)
id_recs$Hab_code <- NULL
}
} # end of repeat for spring migration dates
try({
# Identify the migration start and end dates and distance. Date_times are written in excel format to ensure consistency.
s_migration_start_date <- spring_start
notes_tab[notes_tab$Animal_ID == individs_list[id], ]$SprMig_Start <-
toString(format(s_migration_start_date, format = "%m/%d/%Y %H:%M:%S"))
s_migration_end_date <- spring_end
notes_tab[notes_tab$Animal_ID == individs_list[id], ]$SprMig_End <-
toString(format(s_migration_end_date, format = "%m/%d/%Y %H:%M:%S"))
ss <- id_recs %>%
dplyr::filter(abs(difftime(GMT,s_migration_start_date)) == min(abs(difftime(GMT,s_migration_start_date)))) %>%
dplyr::select(IDTM_X,IDTM_Y) %>%
unique(.)
se <- id_recs %>%
dplyr::filter(abs(difftime(GMT,s_migration_end_date)) == min(abs(difftime(GMT,s_migration_end_date)))) %>%
dplyr::select(IDTM_X,IDTM_Y) %>%
unique(.)
s_migration_distance <- ((ss$IDTM_X - se$IDTM_X)^2 + (ss$IDTM_Y - se$IDTM_Y)^2)^0.5
notes_tab[notes_tab$Animal_ID == individs_list[id], ]$SprMig_Dist <- s_migration_distance
})
}
if(mig == "fa"){
print("Select Fall Migration Dates")
repeat{
# Plot NSD
print(
plt <-ggplot(id_recs,aes(x=GMT,y=Root_NSD)) +
geom_point(color=id_recs$color) + geom_line(color=id_recs$color) +
labs(x="GMT",y="Root NSD",title = paste0("Animal ID: ",individs_list[id]))+
theme_bw()
)
# Zoom in on dates of spring migration
print("Click the earlier date of the range you'd like to zoom to")
windowstart <- ggmap::gglocator(n=1,mercator = F)$GMT
print(plt + geom_vline(xintercept = windowstart))
print("Click the later date of the range you'd like to zoom to")
windowend <- ggmap::gglocator(n=1,mercator = F)$GMT
window_recs <- id_recs[as.numeric(na.omit(id_recs$GMT)) >
windowstart & as.numeric(na.omit(id_recs$GMT)) < windowend, ]
print(plt <- ggplot(window_recs,aes(x=GMT,y=Root_NSD)) +
geom_point(color=window_recs$color) + geom_line(color=window_recs$color) +
labs(x="GMT",y="Root NSD",title = paste0("Animal ID: ",individs_list[id])))
# Pick start/end dates with locator; assign/paste makes new variables for the loop
print("Click the start of the fall migration")
start <- ggmap::gglocator(n=1,mercator = F)$GMT
print(plt <- plt + geom_vline(xintercept = start, color='limegreen'))
print("Click the end of the fall migration")
end <- ggmap::gglocator(n=1,mercator = F)$GMT
print(plt <- plt + geom_vline(xintercept = end, color='darkred'))
# Identify dates in selected range; identify habitat for dates prior to this range
year <- substr(yrs[y], 3, 4)
fall_start <- as.POSIXct(start, origin = "1970-01-01 00:00:00",tz='UTC')
fall_end <- as.POSIXct(end, origin = "1970-01-01 00:00:00",tz='UTC')
if(fall_start > orig_date){ #spring identified in current year sequence
id_recs$habitat <- ifelse(fall_start <= id_recs$GMT & id_recs$GMT <= fall_end,paste0("fallmig", year),
id_recs$habitat)
}else{#spring identified in last year sequence
id_recs$habitat <- ifelse(fall_start <= id_recs$GMT & id_recs$GMT <= fall_end,paste0("fallmig", (as.numeric(year)-1)),
id_recs$habitat)
}
id_recs$Hab_code <- stringr::str_split_fixed(id_recs$habitat,"[:digit:]",2)[,1]
id_recs <- dplyr::left_join(id_recs[,-which(stringr::str_detect(colnames(id_recs),"color")==T)],hab_palette[,c(-1)],by="Hab_code")
# ~~*plotting magic*~~
rmarkdown::render(paste0(scriptdir, "/", "mapbox_plot.Rmd"),
output_file = paste0(individs_list[id], "_graphic.html"),
output_dir = paste0(getwd(), "/unsorted_output/",
herd, "/", yrs[y], sep = ""))
shell.exec(paste0(getwd(), "/unsorted_output/",herd, "/", yrs[y],"/",individs_list[id], "_graphic.html", sep = ""))
if(readline("Open the output file. Look good? Y or N ") == "Y"){
break
}else{
id_recs$habitat <- ifelse(fall_start <= id_recs$GMT & id_recs$GMT <= fall_end,"not assigned",
id_recs$habitat)
id_recs$color <- ifelse(fall_start <= id_recs$GMT & id_recs$GMT <= fall_end,"#000000",
id_recs$color)
id_recs$Hab_code <- NULL
}
} # end of repeat for fall migration dates
try({
if(nrow(id_recs[id_recs$habitat == paste0("fallmig", year), ]) != 0){
# identify the migration start and end dates and distance
f_migration_start_date <- fall_start
notes_tab[notes_tab$Animal_ID == individs_list[id], ]$FallMig_Start <-
toString(format(f_migration_start_date, format = "%m/%d/%Y %H:%M:%S"))
f_migration_end_date <- fall_end
notes_tab[notes_tab$Animal_ID == individs_list[id], ]$FallMig_End <-
toString(format(f_migration_end_date, format = "%m/%d/%Y %H:%M:%S"))
fs <- id_recs %>%
dplyr::filter(abs(difftime(GMT,f_migration_start_date)) == min(abs(difftime(GMT,f_migration_start_date)))) %>%
dplyr::select(IDTM_X,IDTM_Y) %>%
unique(.)
fe <- id_recs %>%
dplyr::filter(abs(difftime(GMT,f_migration_end_date)) == min(abs(difftime(GMT,f_migration_end_date)))) %>%
dplyr::select(IDTM_X,IDTM_Y) %>%
unique(.)
f_migration_distance <- ((fs$IDTM_X - fe$IDTM_X)^2 + (fs$IDTM_Y - fe$IDTM_Y)^2)^0.5
notes_tab[notes_tab$Animal_ID == individs_list[id], ]$FallMig_Dist <- f_migration_distance
}
})
}
if(mig == "wm"){
print("Select Winter Movement Dates")
repeat{
# Plot NSD
print(
plt <-ggplot(id_recs,aes(x=GMT,y=Root_NSD)) +
geom_point(color=id_recs$color) + geom_line(color=id_recs$color) +
labs(x="GMT",y="Root NSD",title = paste0("Animal ID: ",individs_list[id]))+
theme_bw()
)
# Zoom in on dates of spring migration
print("Click the earlier date of the range you'd like to zoom to")
windowstart <- ggmap::gglocator(n=1,mercator = F)$GMT
print(plt + geom_vline(xintercept = windowstart))
print("Click the later date of the range you'd like to zoom to")
windowend <- ggmap::gglocator(n=1,mercator = F)$GMT
window_recs <- id_recs[as.numeric(na.omit(id_recs$GMT)) >
windowstart & as.numeric(na.omit(id_recs$GMT)) < windowend, ]
print(plt <- ggplot(window_recs,aes(x=GMT,y=Root_NSD)) +
geom_point(color=window_recs$color) + geom_line(color=window_recs$color) +
labs(x="GMT",y="Root NSD",title = paste0("Animal ID: ",individs_list[id])))
# Pick start/end dates with locator; assign/paste makes new variables for the loop
print("Click the start of the winter movement")
start <- ggmap::gglocator(n=1,mercator = F)$GMT
print(plt <- plt + geom_vline(xintercept = start, color='limegreen'))
print("Click the end of the winter movement")
end <- ggmap::gglocator(n=1,mercator = F)$GMT
print(plt <- plt + geom_vline(xintercept = end, color='darkred'))
# Identify dates in selected range; identify habitat for dates prior to this range
year <- substr(yrs[y], 3, 4)
s <- as.POSIXct(start, origin = "1970-01-01 00:00:00",tz='UTC')
e <- as.POSIXct(end, origin = "1970-01-01 00:00:00",tz='UTC')
id_recs$habitat <- ifelse(s <= id_recs$GMT & id_recs$GMT <= e,"wntmov",id_recs$habitat)
id_recs$Hab_code <- stringr::str_split_fixed(id_recs$habitat,"[:digit:]",2)[,1]
id_recs <- dplyr::left_join(id_recs[,-which(stringr::str_detect(colnames(id_recs),"color")==T)],hab_palette[,c(-1)],by="Hab_code")
# ~~*plotting magic*~~
rmarkdown::render(paste0(scriptdir, "/", "mapbox_plot.Rmd"),
output_file = paste0(individs_list[id], "_graphic.html"),
output_dir = paste0(getwd(), "/unsorted_output/",
herd, "/", yrs[y], sep = ""))
shell.exec(paste0(getwd(), "/unsorted_output/",herd, "/", yrs[y],"/",individs_list[id], "_graphic.html", sep = ""))
if(readline("Open the output file. Look good? Y or N ") == "Y"){
break
}else{
id_recs$habitat <- ifelse(s <= id_recs$GMT & id_recs$GMT <= e,"not assigned",
id_recs$habitat)
id_recs$color <- ifelse(s <= id_recs$GMT & id_recs$GMT <= e,"#000000",
id_recs$color)
id_recs$Hab_code <- NULL
}
} # end of repeat for winter movement dates
}
if(mig == "sm"){
print("Select Summer Movement Dates")
repeat{
# Plot NSD
print(
plt <-ggplot(id_recs,aes(x=GMT,y=Root_NSD)) +
geom_point(color=id_recs$color) + geom_line(color=id_recs$color) +
labs(x="GMT",y="Root NSD",title = paste0("Animal ID: ",individs_list[id]))+
theme_bw()
)
# Zoom in on dates of spring migration
print("Click the earlier date of the range you'd like to zoom to")
windowstart <- ggmap::gglocator(n=1,mercator = F)$GMT
print(plt + geom_vline(xintercept = windowstart))
print("Click the later date of the range you'd like to zoom to")
windowend <- ggmap::gglocator(n=1,mercator = F)$GMT
window_recs <- id_recs[as.numeric(na.omit(id_recs$GMT)) >
windowstart & as.numeric(na.omit(id_recs$GMT)) < windowend, ]
print(plt <- ggplot(window_recs,aes(x=GMT,y=Root_NSD)) +
geom_point(color=window_recs$color) + geom_line(color=window_recs$color) +
labs(x="GMT",y="Root NSD",title = paste0("Animal ID: ",individs_list[id])))
# Pick start/end dates with locator; assign/paste makes new variables for the loop
print("Click the start of the summer movement")
start <- ggmap::gglocator(n=1,mercator = F)$GMT
print(plt <- plt + geom_vline(xintercept = start, color='limegreen'))
print("Click the end of the summer movement")
end <- ggmap::gglocator(n=1,mercator = F)$GMT
print(plt <- plt + geom_vline(xintercept = end, color='darkred'))
# Identify dates in selected range; identify habitat for dates prior to this range
year <- substr(yrs[y], 3, 4)
s <- as.POSIXct(start, origin = "1970-01-01 00:00:00",tz='UTC')
e <- as.POSIXct(end, origin = "1970-01-01 00:00:00",tz='UTC')
id_recs$habitat <- ifelse(s <= id_recs$GMT & id_recs$GMT <= e,"summov",id_recs$habitat)
id_recs$Hab_code <- stringr::str_split_fixed(id_recs$habitat,"[:digit:]",2)[,1]
id_recs <- dplyr::left_join(id_recs[,-which(stringr::str_detect(colnames(id_recs),"color")==T)],hab_palette[,c(-1)],by="Hab_code")
# ~~*plotting magic*~~
rmarkdown::render(paste0(scriptdir, "/", "mapbox_plot.Rmd"),
output_file = paste0(individs_list[id], "_graphic.html"),
output_dir = paste0(getwd(), "/unsorted_output/",
herd, "/", yrs[y], sep = ""))
shell.exec(paste0(getwd(), "/unsorted_output/",herd, "/", yrs[y],"/",individs_list[id], "_graphic.html", sep = ""))
if(readline("Open the output file. Look good? Y or N ") == "Y"){
break
}else{
id_recs$habitat <- ifelse(s <= id_recs$GMT & id_recs$GMT <= e,"not assigned",
id_recs$habitat)
id_recs$color <- ifelse(s <= id_recs$GMT & id_recs$GMT <= e,"#000000",
id_recs$color)
id_recs$Hab_code <- NULL
}
} # end of repeat for summer movement dates
}
BR <- readline("Classify more movements? Y or N ")
if(BR == "N"){
notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Strategy <- readline("Identify final migration strategy.
Please list M (migratory), MM(mixed-migratory), or U(unclear) ")
break
}
} # end of repeat for migration date classifications
} # end of Date Selection
rmarkdown::render(paste0(scriptdir, "/", "mapbox_plot.Rmd"),
output_file = paste0(individs_list[id], "_graphic.html"),
output_dir = paste0(getwd(), "/unsorted_output/",
herd, "/", yrs[y], sep = ""))
shell.exec(paste0(getwd(), "/unsorted_output/",herd, "/", yrs[y],"/",individs_list[id], "_graphic.html", sep = ""))
if(readline("Open the output file. Look good? Y or N ") == "Y"){break}
} #classify repeat
# Finish Classifying Dates
if(exists("discard_all")){
discard_all$habitat <- "Discard"
discard_all$Hab_code <- "Discard"
discard_all <- dplyr::left_join(discard_all[,-which(stringr::str_detect(colnames(discard_all),"color")==T)],hab_palette[,c(-1)],by="Hab_code")
id_recs <- id_recs %>%
rbind(.,discard_all) %>%
dplyr::arrange(.,GMT)
rm(discard_all,discard,discarded)
}
id_recs <- classify_dates(id_recs)
#~~*plotting magic*~~
rmarkdown::render(paste0(scriptdir, "/", "mapbox_plot.Rmd"),
output_file = paste0(individs_list[id], "_graphic.html"),
output_dir = paste0(getwd(), "/unsorted_output/",
herd, "/", yrs[y], sep = ""))
shell.exec(paste0(getwd(), "/unsorted_output/",herd, "/", yrs[y],"/",individs_list[id], "_graphic.html", sep = ""))
if(readline("Look good? Y or N ") == "Y"){break}
}
if(notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Skip == "Y"){break}
} # end of repeat for individual
notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Revisit <- readline("Revisit this animal later? Y or N ")
notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Notes <- readline("Write whatever notes you want ")
# Paste labels onto the graphics
if(notes_tab[notes_tab$Animal_ID == individs_list[id], ]$Skip == "Y"){
id_recs$habitat <- "not assigned"
notes_tab[notes_tab$Animal_ID == individs_list[id], !names(notes_tab) %in% c("Skip", "Months_of_Data", "Animal_ID", "Notes","Region","AnchorDate","Revisit")] <- NA # ; wipe out any data created for this animal, b/c skipping it now
}
# Flag Migrations that might be too short (less than 5 km)
notes_tab[notes_tab$Animal_ID == individs_list[id],]$FlagMig <- ifelse(is.na(notes_tab[notes_tab$Animal_ID == individs_list[id],]$SprMig_Dist)==T & is.na(notes_tab[notes_tab$Animal_ID == individs_list[id],]$FallMig_Dist)==T, F,
ifelse(is.na(notes_tab[notes_tab$Animal_ID == individs_list[id],]$FallMig_Dist)==T & is.na(notes_tab[notes_tab$Animal_ID == individs_list[id],]$SprMig_Dist)==F & notes_tab[notes_tab$Animal_ID == individs_list[id],]$SprMig_Dist <= 5000, T,
ifelse(is.na(notes_tab[notes_tab$Animal_ID == individs_list[id],]$FallMig_Dist)==F & is.na(notes_tab[notes_tab$Animal_ID == individs_list[id],]$SprMig_Dist)==T & notes_tab[notes_tab$Animal_ID == individs_list[id],]$FallMig_Dist <= 5000, T,
ifelse(notes_tab[notes_tab$Animal_ID == individs_list[id],]$FallMig_Dist <= 5000 | notes_tab[notes_tab$Animal_ID == individs_list[id],]$SprMig_Dist <= 5000, T, F))))
print("Finalizing output graphics.")
id_recs <- id_recs[year(id_recs$GMT) == yrs[y], ] # redefine id_records, now that id_recs is classified
rmarkdown::render(paste0(scriptdir, "/", "mapbox_plot.Rmd"),
output_file = paste0(individs_list[id], "_graphic.html"),
output_dir = paste0(getwd(), "/unsorted_output/", herd,
"/", yrs[y], sep = ""))
print("Writing output tables.")
fwrite(id_recs, paste0(getwd(), "/unsorted_output/", herd, "/", yrs[y],
"/", individs_list[id], ".csv"), row.names = FALSE,
dateTimeAs = c("write.csv"))
fwrite(notes_tab, paste0(getwd(), "/unsorted_output/", herd, "/", yrs[y],
"/", yrs[y], "_overall.csv"), row.names = FALSE,
dateTimeAs = c("write.csv"))
print("Updating overall table with new values.")
all_recs <- all_recs %>%
dplyr::anti_join(.,id_recs[,-which(stringr::str_detect(colnames(id_recs),"color|habitat|Hab_code")==T)],by=c("Animal_ID","GMT")) %>%
rbind(.,id_recs[,-which(stringr::str_detect(colnames(id_recs),"Hab_code")==T)]) %>%
dplyr::arrange(.,Animal_ID,GMT)
savecounter <- savecounter + 1
if(savecounter == 5){
print("5 animals done. Backing up the data table.")
fwrite(all_recs, outfile, row.names = FALSE, dateTimeAs = c("write.csv"))
savecounter <- 0
} else if (readline("Save manually? Do this if you are exiting the script. Y or N ") == "Y") {
fwrite(all_recs, outfile, row.names = FALSE, dateTimeAs = c("write.csv"))
savecounter <- 0
} else {
print(paste("Overall file will back up automatically after", 5 - savecounter, "more animals"))
}
grDevices::dev.off()
system2("taskkill",args="/im chrome.exe")
} # individual loop
} # year loopMigrations
I am currently an Associate Research Scientist for the Wyoming Cooperative Fish and Wildlife Research Unit modeling ungulate seasonal ranges and migrations for Idaho Department of Fish and Game in support of S.O. 3362. One of my primary duties is to classify migrations of Idaho’s mule deer, elk, and pronghorn using net-squared displacement analysis. I also maintain and enhance the analysis code to streamline the workflow and allow for flexibility to accommodate complex migration patterns.
Net-squared displacement analysis