Mule deer
300,081 winter range locations
987,558 total locations
22 attributes
Elk
764,119 winter range locations
2,661,400 total locations
19 attributes
Pronghorn
187,860 winter range locations
731,025 total locations
19 attributes
Mule deer
300,081 winter range locations
987,558 total locations
22 attributes
Elk
764,119 winter range locations
2,661,400 total locations
19 attributes
Pronghorn
187,860 winter range locations
731,025 total locations
19 attributes
I am currently an Associate Research Scientist for the Wyoming Cooperative Fish and Wildlife Research Unit modeling ungulate seasonal ranges and migrations for Idaho Department of Fish and Game in support of S.O. 3362. So far, I’ve completed winter range models for mule deer using the random forest machine learning algorithm in a resource selection function framework. I assembled these predictions into a dashboard to assist in assessing the predictions and outputs. Included in the dashboard are maps depicting the average predicted winter range for mule deer in each of IDFG’s mule deer data analysis units (DAUs). In addition, I’ve started to analyze the changes in winter range predictions across years.
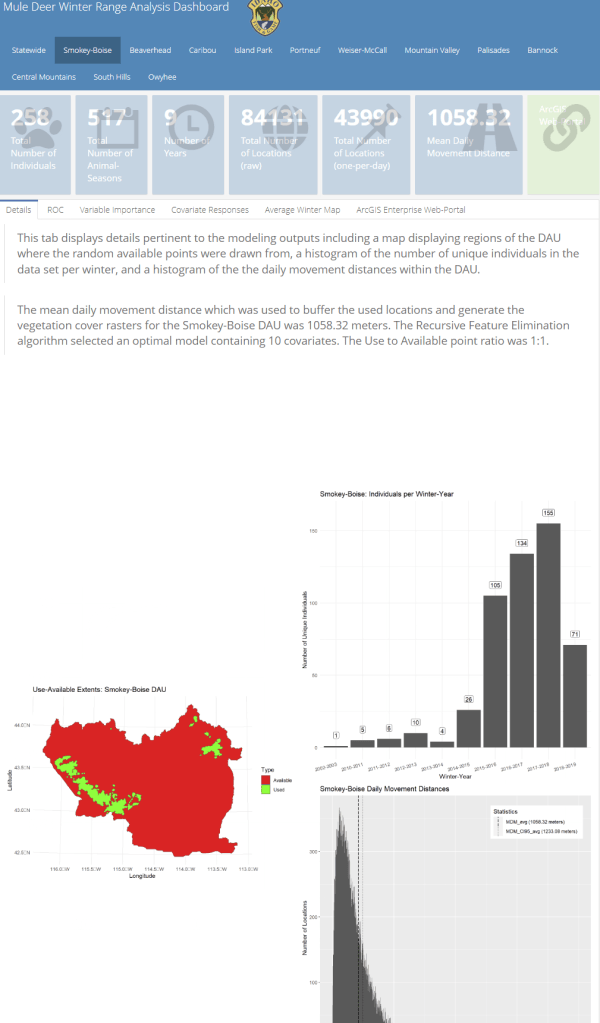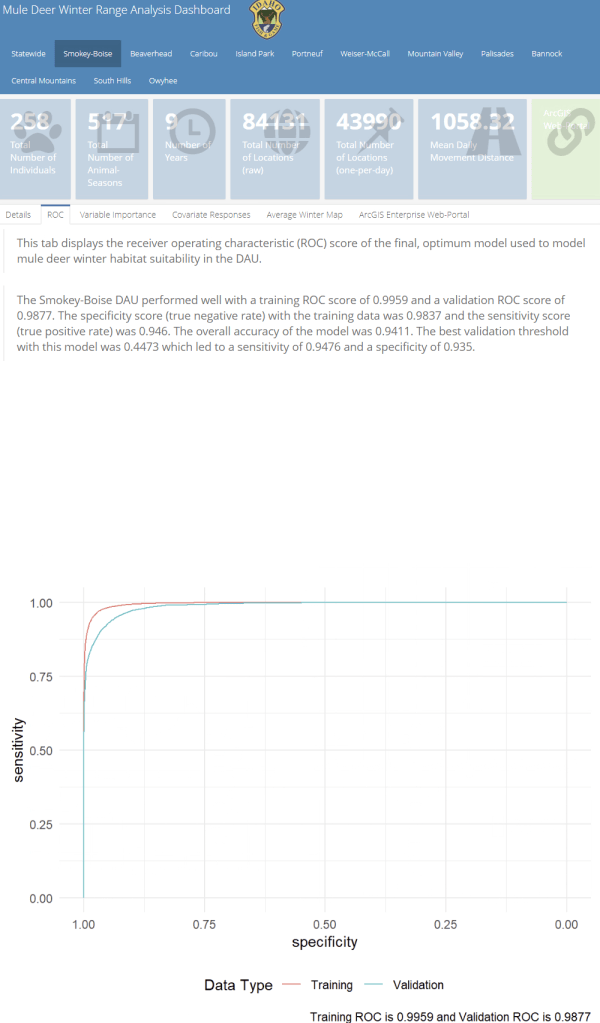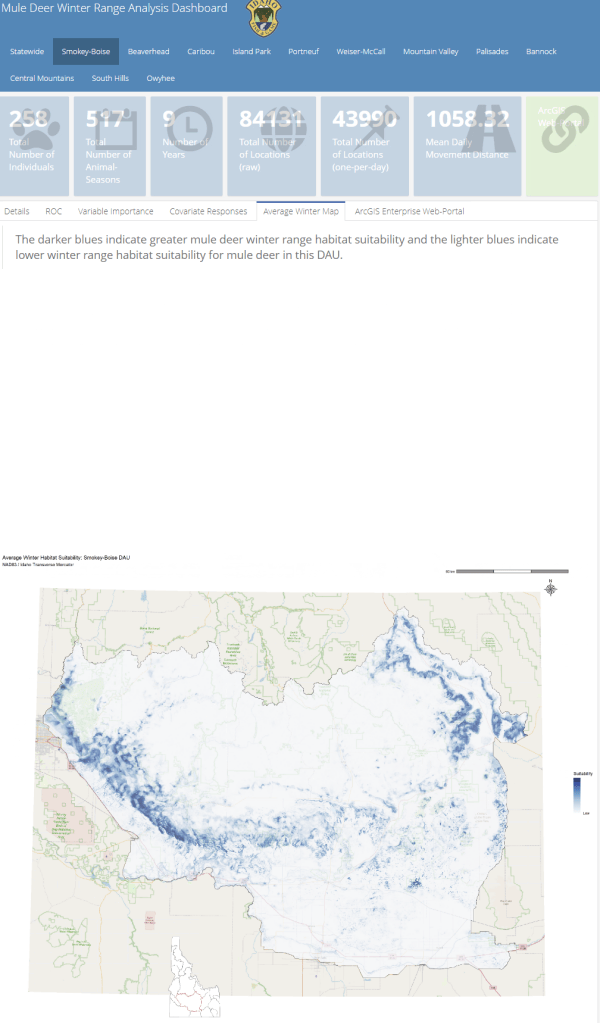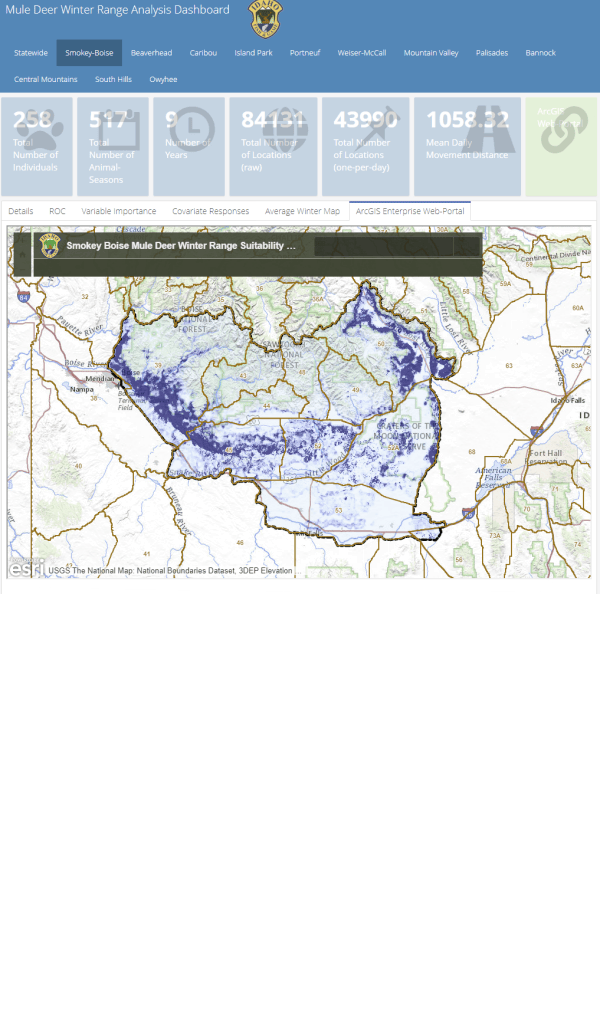
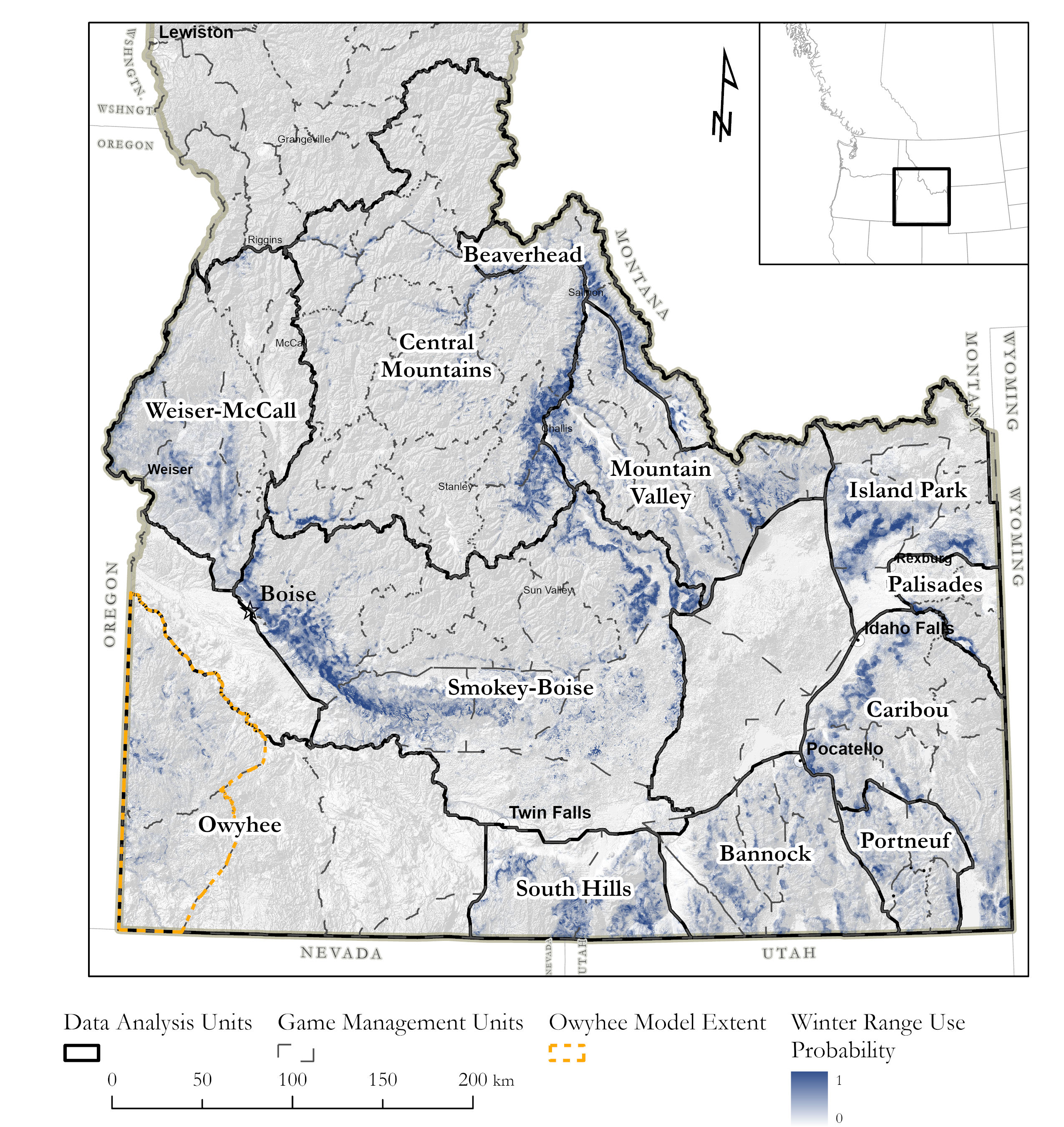
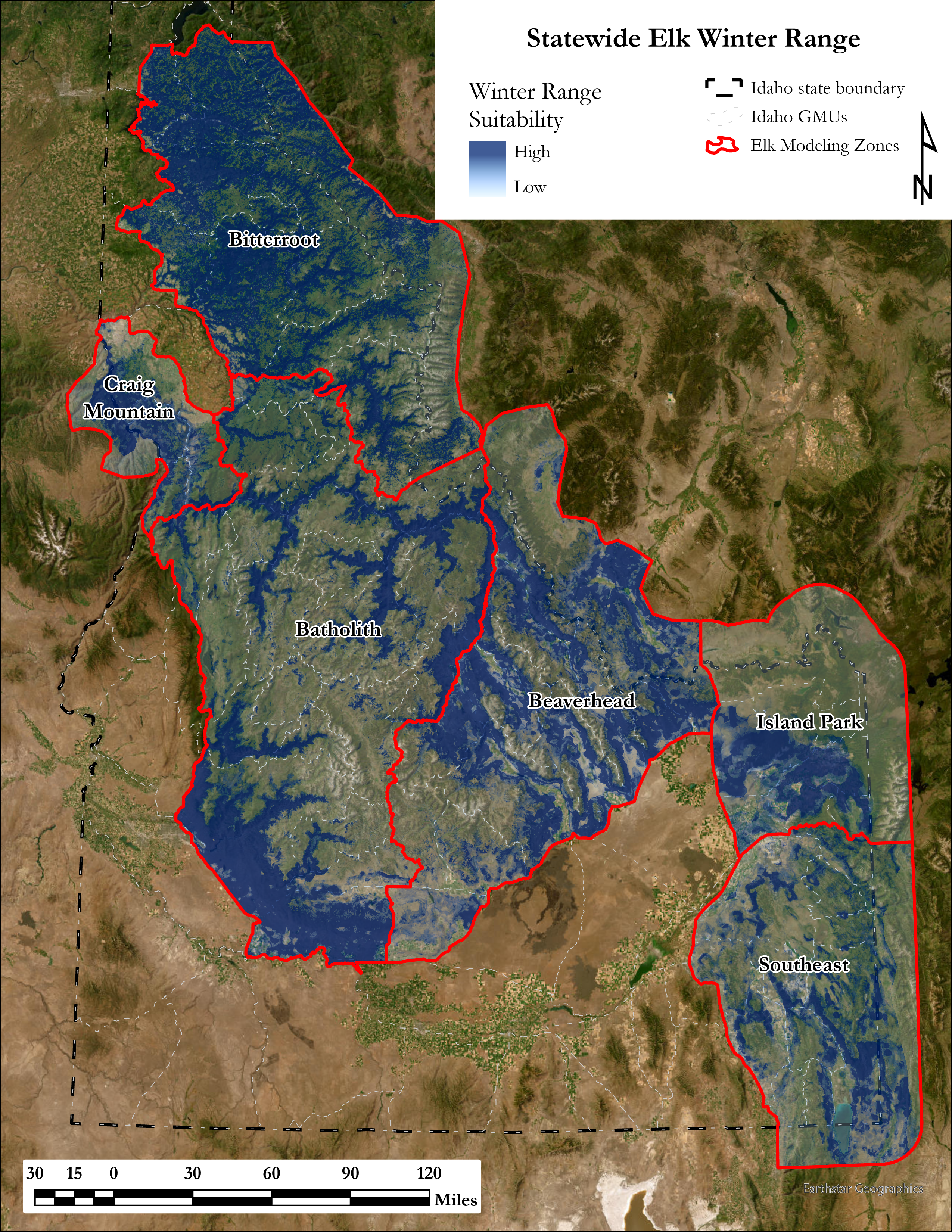
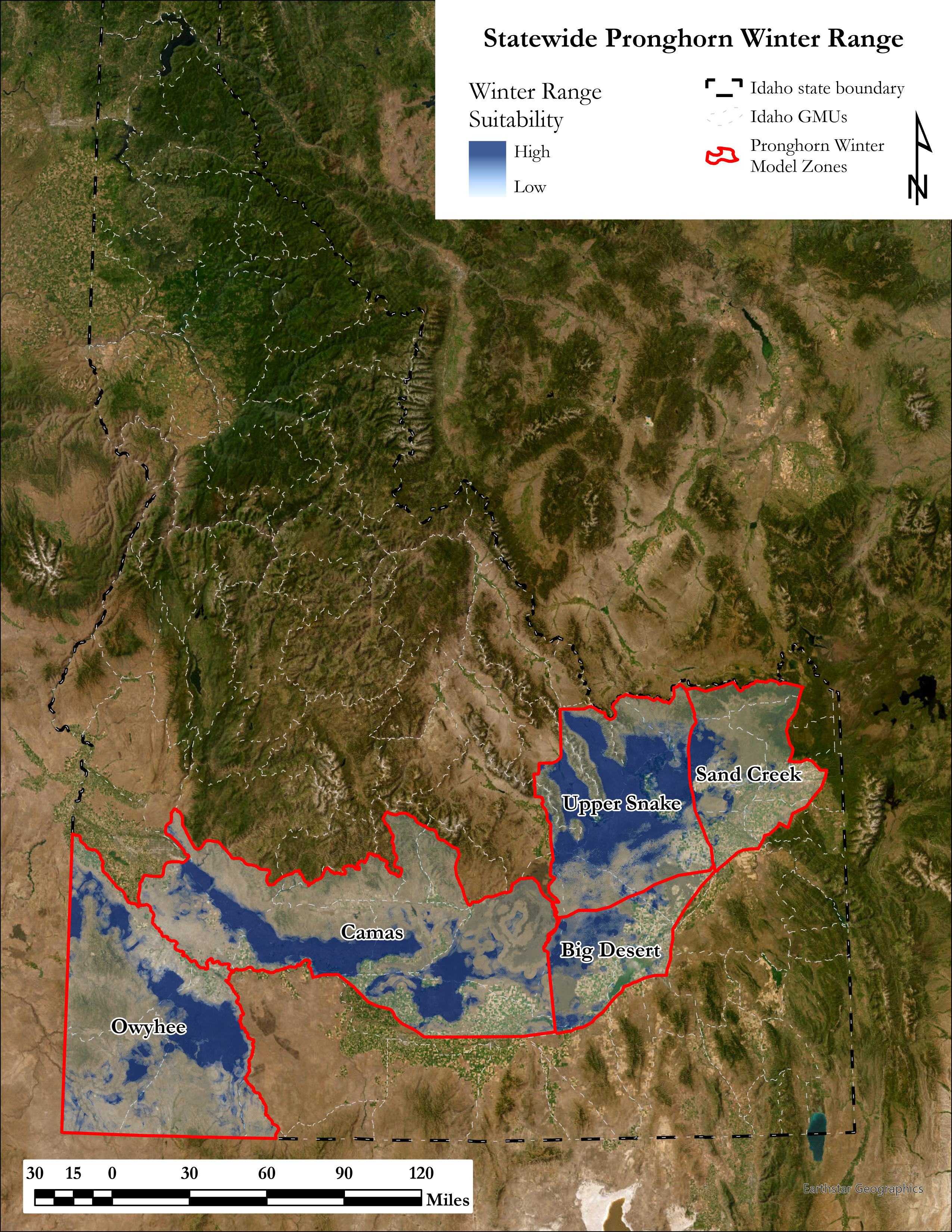
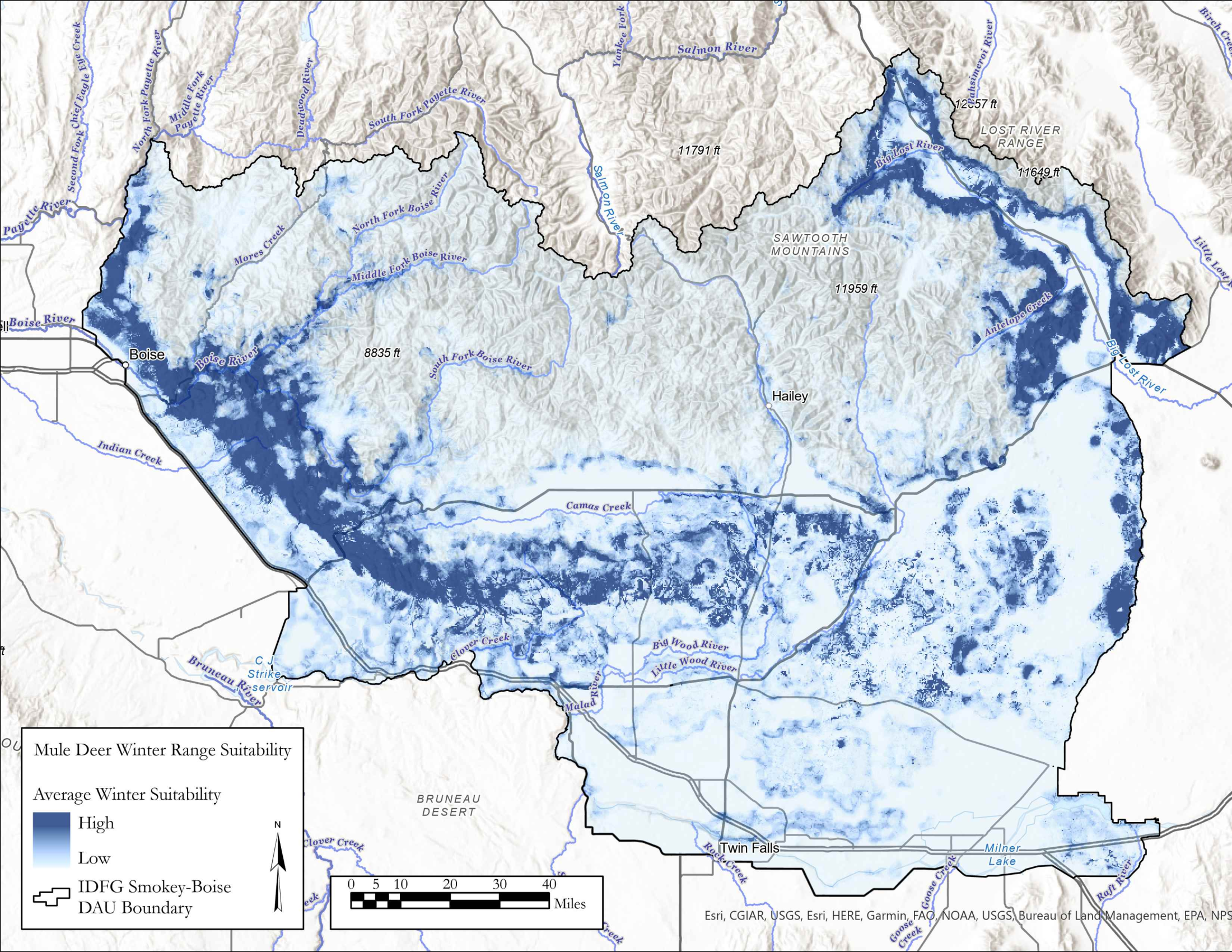
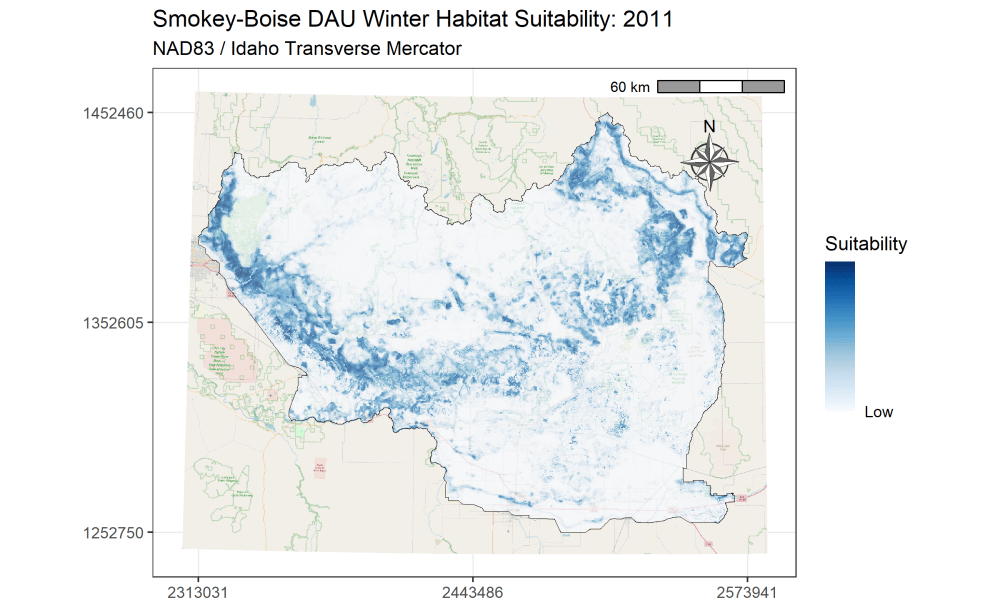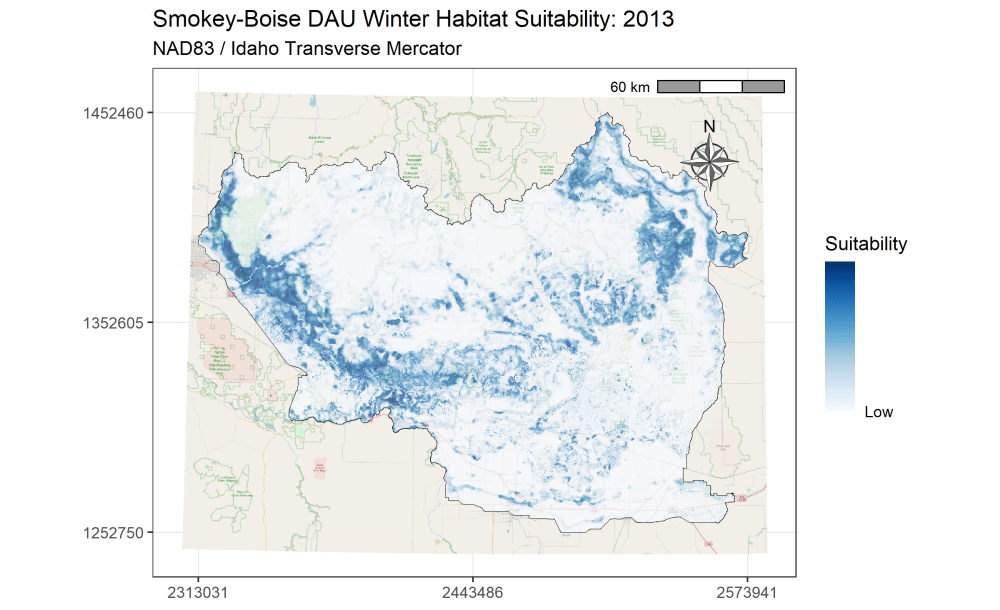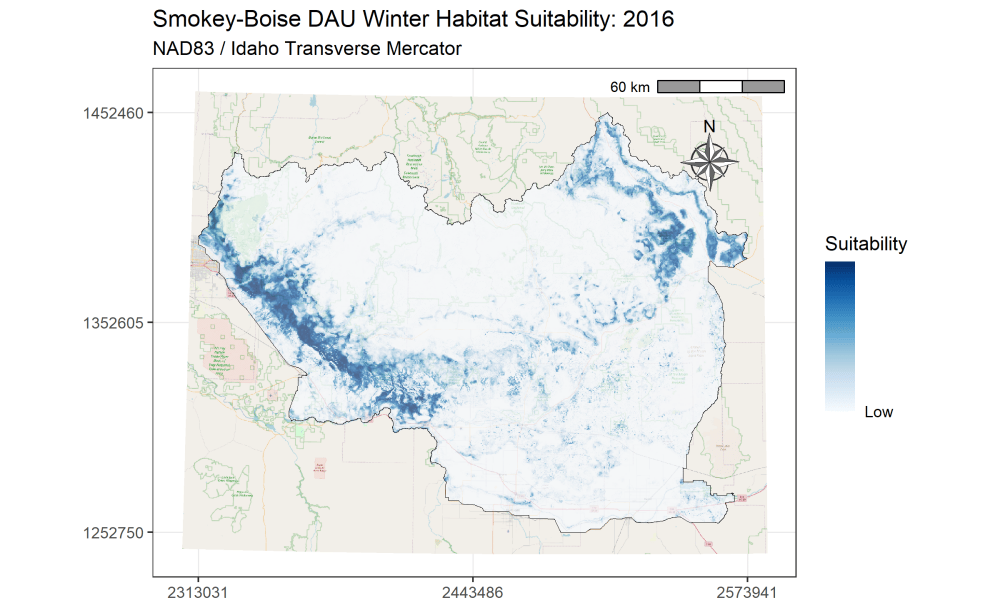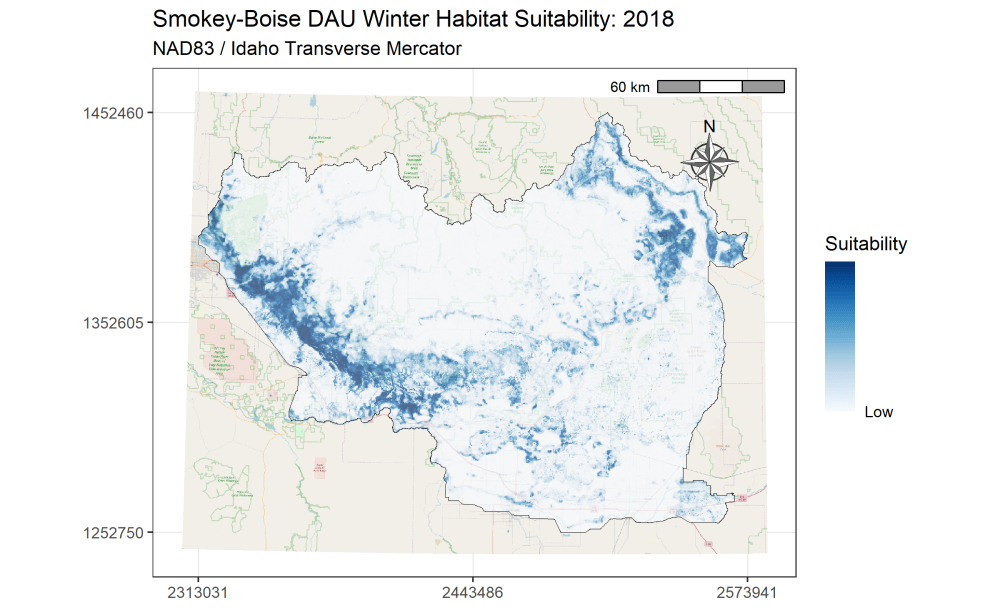
A major component of the seasonal range models is IDFG’s vegetation classification covariates which are derived from LANDFIRE. The models explicitly require each vegetation class to be transformed to a focal value based on the average daily movement distance of individuals in the DAU. To streamline this covariate transformation, I wrote a Python function using ESRI’s arcpy and Spatial Analyst tools, which are called by an R script I wrote to iterate the process over each collection of DAU covariates.
# --------------------------------------------------------
# Calculate Hurley Vegetation Focal Statistics
# Author: Robert Ritson, Associate Research Scientist (UW)
# Last Updated: 5/10/2022
# Description: Calculate square meters of Hurley
# vegetation classes (binary rasters derrived from LANDFIRE)
# given a mean daily movement distance (from an ungulate population),
# clipped by a population DAU boundary
# --------------------------------------------------------
#Import modules
import arcpy
from arcpy.sa import *
import datetime
import csv
import os
arcpy.CheckOutExtension("Spatial")
#Define function
def hveg_focal(path_to_dau, mdm_ci95, path_to_hveg, dau_hveg_folder, scratch = "C:\\Users\\rritson\\Documents\\ArcGIS\\scratch\\"):
try:
print 'begin script on '+datetime.datetime.now().date().isoformat()+' at '+datetime.datetime.now().time().isoformat()[0:8]
# Set path to folders and workspace
dauws = path_to_dau
rastws = path_to_hveg
outws = dau_hveg_folder
neighborhood = arcpy.sa.NbrCircle(mdm_ci95,"MAP")
arcpy.env.overwriteOuptput = False
arcpy.env.workspace = scratch
# Buffer Selected DAU by provided mean daily movement
print 'buffering DAU shape by mean daily movement input (meters)'
arcpy.Buffer_analysis(dauws,scratch+'\\DAU_Buffer.shp', mdm_ci95,"FULL","ROUND","NONE","","PLANAR")
# List Hurley Vegetation Rasters
arcpy.env.workspace = rastws
try:
print 'listing Hveg rasters'
hveg_list = [hveg for hveg in arcpy.ListRasters()]
except: print arcpy.GetMessages(2)
# Clip Hurley Vegetation Raster to buffered DAU and calculate focal statistics
arcpy.env.workspace = scratch
for hveg in hveg_list:
print 'loading raster '+hveg
arcpy.MakeRasterLayer_management(rastws+'/'+hveg,'temp_rast.tif',"",scratch+'/DAU_Buffer.shp',"") #load raster and clip to buffered DAU extent
print 'calculating focal statistics'
arcpy.CheckOutExtension("Spatial")
fstat = FocalStatistics('temp_rast.tif', neighborhood, "SUM", "DATA") #Focal sum of binaries by mean daily movement distance
print 'converting to square meters'
outrast = fstat * (30^2) #multiply focal sum by cell dimensions (convert to square meters)
outrast.save(outws+hveg) #save output
print hveg+' raster saved to '+outws+' at '+datetime.datetime.now().time().isoformat()[0:8]
print 'Finished: Completed Hurley vegetation density rasters for '+dauws+' located in '+outws
print 'script completed on '+datetime.datetime.now().date().isoformat()+' at '+datetime.datetime.now().time().isoformat()[0:8]
except: print arcpy.GetMessages(2)
arcpy.CheckInExtension("Spatial")
# # # # # END OF FUNCTION # # # # # # ### Hurley Vegetation Density Raster Calculations ##
### Loop through remaining DAUs (~10min per DAU) ###
#Load package
require(dplyr)
#devtools::install_github("rstudio/reticulate")
#Sys.setenv(RETICULATE_PYTHON = "C:\\Python27\\ArcGIS10.8\\\\pythonw.exe")
Sys.setenv(RETICULATE_PYTHON = 'C:/Users/rritson/AppData/Local/Programs/ArcGIS/Pro/bin/Python/envs/arcgispro-py3/python.exe')
require(reticulate)
#DAU List
dau_list <- sf::st_read('C:/Users/rritson/Documents/Projects/MuleDeer_SeasonalRanges/DAU_BBox_10kbuff.shp') %>%
as.data.frame(.) %>%
dplyr::select(NAME) %>%
#dplyr::filter(NAME == "Island Park")
dplyr::filter(NAME %in% c("Bannock","Caribou","Mountain Valley","Portneuf","Weiser-McCall"))
#dplyr::filter(!(NAME %in% c("Bitterroot","Panhandle","Lower Salmon","Snake River","Smokey-Boise","Island Park")))
#DAU shape (all)
dau_shp <- sf::st_read('C:/Users/rritson/Documents/Projects/MuleDeer_SeasonalRanges/DAU_BBox_10kbuff.shp') %>%
#dplyr::select(-GlobalID,-Shape_Leng,-Shape_Area) %>%
sfheaders::sf_remove_holes(.)
#Movement Data
dat <- readRDS("C:/Users/rritson/Documents/Projects/MuleDeer_SeasonalRanges/Winter_RSF/MD_Winter_Locs_1pd.rds") %>% as.data.frame(.)
# Check R Session Info
if(sessionInfo()$R.version$arch != "i386"){
print("STOP: Must run R in 32-bit Architecture to source 'arcpy'")
}
if(paste0(sessionInfo()$R.version$major,".",sessionInfo()$R.version$minor) > "4.0.4"){
print("STOP: Must run R in version prior or equal to 4.0.4 to source 'arcpy'")
# reticulate does not want to work with R version 4.1.0...
}
# Initiate Python before beginning loop
#reticulate::use_python("C:\\Python27\\ArcGIS10.8\\\\pythonw.exe", required = T)
reticulate::use_python('C:/Users/rritson/AppData/Local/Programs/ArcGIS/Pro/bin/Python/envs/arcgispro-py3/python.exe', required = T)
reticulate::py_config()
reticulate::source_python("C:/Users/rritson/Documents/Python Scripts/hveg_focal_func.py")
#reticulate::source_python("C:/Users/rritson/Documents/Python Scripts/hveg_focal_func_sing.py") #for dealing with a single raster (must change file paths below if so)
# Loop through DAUs
for (i in 1:nrow(dau_list)){
# Select DAU
print(paste("Beginning DAU:",dau_list[i,],"(",nrow(dau_list)-i,"remaining)..."))
dau_sel <- dau_list[i,]
# Create folder for outputs
#print("Creating output folder...")
#dir.create("F:/Seasonal_range_covars/mdmci95_2buff")
#dir.create(paste0("F:/Seasonal_range_covars/mdmci95_2buff/",dau_sel))
# Write DAU shapefile
#print("Writing shapefile...")
#dau <- dau_shp %>% dplyr::filter(NAME == dau_sel)
#sf::write_sf(dau,paste0('C:/Users/rritson/Documents/Projects/MuleDeer_SeasonalRanges/SeasonalRanges/',dau_sel,".shp"),append=F)
# Get Mean Daily Movements for DAU
print("Accessing mean daily movement (95% CI, in meters)...")
mdm_ci95 <- dat %>% dplyr::filter(DAU == dau_sel) %>% dplyr::select(MDM_CI95_avg) %>% dplyr::slice(1)
mdm_ci95 <- mdm_ci95[[1]]
mdm_ci95_2 <- mdm_ci95 / 2
# Calculate Hurley vegetation density raster for DAU: Mean Daily Movement
print("Reticulating HVeg Density Raster Calculation (Mean Daily Movement)...")
hveg_focal(path_to_dau = paste0('C:/Users/rritson/Documents/Projects/MuleDeer_SeasonalRanges/SeasonalRanges/',dau_sel,".shp"),
mdm_ci95 = mdm_ci95, #mean daily movement
path_to_hveg = "F:\\Seasonal_range_covars\\hveg",
dau_hveg_folder = paste0("F:/Seasonal_range_covars/mdmci95_buff/",dau_sel,"/"), #CHANGE THIS!!! (to match output folder)
scratch = "C:/Users/rritson/Documents/ArcGIS/scratch")
#Delete temp files
lapply(list.files("C:/Users/rritson/Documents/ArcGIS/scratch",full.names = T),file.remove)
gc()
# Calculate Hurley vegetation density raster for DAU: One-half Mean Daily Movement
print("Reticulating HVeg Density Raster Calculation (One-Half Mean Daily Movement)...")
hveg_focal(path_to_dau = paste0('C:/Users/rritson/Documents/Projects/MuleDeer_SeasonalRanges/SeasonalRanges/',dau_sel,".shp"),
mdm_ci95 = mdm_ci95_2, #one-half mean daily movement
path_to_hveg = "F:\\Seasonal_range_covars\\hveg",
dau_hveg_folder = paste0("F:/Seasonal_range_covars/mdmci95_2buff/",dau_sel,"/"), #CHANGE THIS!!! (to match output folder)
scratch = "C:/Users/rritson/Documents/ArcGIS/scratch")
#Delete temp files
lapply(list.files("C:/Users/rritson/Documents/ArcGIS/scratch",full.names = T),file.remove)
gc()
}